Series Overview: Hi, my name is Shari and like the majority of our readers I don’t have a degree in science. What I do have is almost a decade of experience working closely with Genetic Counselors, and a passion for sharing genomic information in an approachable and easy to understand way. This passion was developed early in my career, fueled by my quickly building frustration about the over-complicated resources I found during initial attempts at self-guided learning. Genomics 101 is a blog series by GenomicMD that aims to be a solution to that frustration for others. By breaking down the complex language surrounding this field, we hope to empower people (like you!) to be more informed about how genomics can affect their healthcare journey.
Welcome back to Genomics 101, the blog series where we decode the intricate world of genetics into easy and understandable language. In our last Gen101 post, we explored different types of genetic inheritance, from monogenic to polygenic traits. Today, we will examine the world of Polygenic Risk Scores (PRS), a groundbreaking tool that represents a more comprehensive approach to understanding the genetics of multifactorial medical conditions. Keep reading to learn more about how this new twist on an established technology is transforming personalized healthcare.
Before we take our dive into PRS, let's quickly recap polygenic inheritance, specifically in relation to personal health. As mentioned in our last blog: unlike monogenic traits (which are governed by single genes) or oligogenic traits (which involve a few cooperating genes), polygenic traits are the result of complex interactions between multiple genes, variants, and environmental factors. Think of them like a mosaic of genetic and environmental information that contribute to a variety of personal qualities such as our height and skin color - or sometimes even our risk for certain health conditions like obesity, diabetes, and heart disease.

Now that we’ve reviewed what they ARE, let’s move on to discussing HOW scientists screen for polygenic traits. Before creating any type of polygenic test, scientists must first analyze vast amounts of genetic data from large populations. This data usually comes from Genome-Wide Association Studies (GWAS). GWAS are a well established method of genetic research that involves scanning the DNA of a large population (sometimes up to millions of people!) to identify genetic variants that are associated with specific traits or diseases. These genetic variants are often referred to as Single Nucleotide Polymorphisms (SNPs), which is just a fancy name for very common and VERY small genetic changes that everyone has in their DNA. If you recall our blog post “A brief introduction to DNA”, we defined nucleotides as the chemical compounds that sit on opposite sides of the ‘double helix’ and form bonds with one another like rungs in a ladder. This means that when we describe SNPs, we are saying these genetic changes are so small that they only occur at ONE nucleotide placement in our DNA. Because of their size and relatively common occurrence, most SNPs (outside of some pretty rare and well-known monogenic variants which are known to be associated with more significant disease risks) are benign, meaning that they are not known to contribute to any disease. This doesn’t mean that ALL Single Nucleotide Polymorphisms are unimportant to our health, though. Falling between these two extremes sit polygenic SNPs. Polygenic SNPs typically have a mild to moderate disease risk by themselves, but when researchers add them together they are able to collectively give us information about our personal likelihood of developing certain conditions. GWAS studies use these SNPs as tiny guideposts in our genetic map, leading researchers to clues that pave the way for advancements in medicine like new methods of screening, treatment, and prevention.

Laboratories (like GenomicMD) use a method that combines information about multiple SNPs that have small but clinically established effects to generate something called a Polygenic Risk Score (PRS), which is a tool used to assess a person's risk of developing a given condition or disease and compare it to that of the average population. So essentially, instead of asking a ‘yes’ or ‘no’ question like the majority of genetic testing methods (Do you have this specific genetic variant or not?), PRS are instead asking “how much” risk a person has compared to the average person by first asking “how many” of these particular SNPs they have. When referencing a Polygenic Risk Score, a higher score indicates a greater genetic risk as compared to the average population, while a lower score (as I’m sure you’ve guessed) suggests a lower risk. For example, a PRS of 1.0 would mean a person is at average genomic risk - meaning they are just as likely as anyone else to develop this condition based on their DNA alone. Alternatively, a score that is below 1.0 would mean this person has a lower than average genetic risk, and one that is 2.0 would indicate they are twice as likely to develop this particular health issue when compared to the average person.
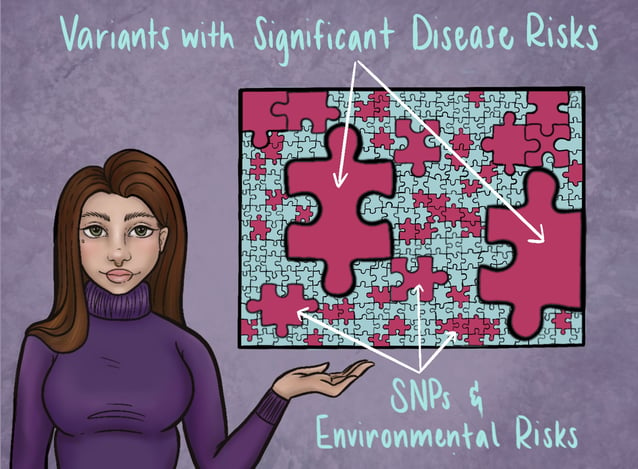
Think of a person’s total risk to develop a given disease or condition like a puzzle. Some puzzles are very simple and only require a few pieces to complete, while others are much more intricate and contain pieces of varying shapes and sizes. Puzzles with only a few pieces are like monogenic or oligogenic disease, where there are only a few factors that contribute to their development. On the other hand, a puzzle representing polygenic disease would be much more complex. It may have a couple of large pieces that take up a significant chunk of space, but it would also have many connecting pieces of varying sizes that surround them and tie everything together. In this analogy, the larger puzzle pieces can be compared to those genetic variants which are linked to significant disease risks, and the smaller pieces are more like environmental risk factors and those SNPs we mentioned earlier. The key takeaway here is that each piece of the puzzle, no matter how small, represents important information that contributes to the complete picture of a person’s overall disease risk. The use of risk assessment tools like Polygenic Risk Scores can help healthcare teams highlight often-overlooked genetic information, allowing them to more easily see the ‘bigger picture’.
With all of this being said, it's essential to keep in mind that though this technology can provide a lot of context to conversations about disease risk, PRS (like any tool for risk assessment) is not a crystal ball. It cannot offer an ABSOLUTE prediction about whether you will or won't develop a certain condition. At this time there is no test that can definitively predict whether a person will develop these sorts of multifactorial conditions. Because of this, it is best to focus on what science CAN provide. Polygenic Risk Scores can offer valuable information about an individual's genetic risk for medical conditions that are often very actionable, meaning there are established steps people can take under the guidance of their clinical team that lead to better healthcare outcomes. Clinicians can use PRS alone or alongside other risk stratification tools, like family health histories and other laboratory tests, to guide discussions about lifestyle changes, early screening, and preventive measures tailored to an individual's genetic risk profile and personal needs.
In conclusion, Polygenic Risk Scores are an innovative tool that utilize well-established methods of research to create a more comprehensive approach to genomic medicine. We at GenomicMD truly believe that the polygenic approach to health screening embraces the beauty and complexity of our individual genetic makeup – one which allows our team the opportunity to offer a unique and clinically precise glimpse into the future of medicine. Our hope is that by giving insight into genetic predispositions both great and small, we can make understanding a patient’s inherent genetic risk simple, allowing healthcare teams to focus on suggesting and creating treatments and interventions that can be adjusted to the needs of each individual. Thank you for coming with us on this voyage through the science of polygenic risk scores - we hope you join us again for our next installment of Genomic101!
Blog Glossary:
GWAS - A widely-used method of genetic research that involves scanning the DNA of large populations (sometimes up to millions of people!) to identify genetic variants that are associated with specific traits or diseases
Polygenic risk score - calculations that sum up the effects of many genetic variants in order to determine an individual’s risk of developing a certain disease
Single Nucleotide Polymorphism (SNPs) - common genetic variations that are small changes in the genome created by the replacement of a nucleotide for another
Benign - Something that is not believed to be disease-causing
Risk Stratification Tools - clinical approaches that healthcare teams use, like family health histories and laboratory tests, to guide discussions about actions that can be taken to reduce the risk of disease
Actionable - When a disease or condition is has established steps or guidelines that people can take under the counsel of their healthcare team which typically lead to better healthcare outcomes